Discovering a Novel Therapy for Urinary Tract Infections Through Outsourcing In-silico Drug Discovery
Antimicrobial resistance: The return of an old nemesis
Antimicrobial therapies are one of the great human achievements of the 20th century. For centuries, bacterial infection was the biggest killer of people worldwide, with infamous diseases such as tuberculosis and the bubonic plague seemingly unstoppable. If a person caught a serious bacterial infection, then the prognosis was bleak, however, in the 1940’s, scientists turned the table on these invisible killers, with the discovery of the first antibiotic, penicillin. Today, as well as bacterial; viral, fungal and parasitic infections are all treatable with small molecule antimicrobial therapies obtained from a local pharmacy, within a couple of hours of feeling a slight cough or high temperature. Surely then, we’ve turned our back on the days where minor infection could pose a threat to life? If antimicrobial resistance (AMR) continues, we may soon be back in the dark ages of medicine, where superbugs are able to survive our best defenses.
AMR was first recognized by the UK government in 2014, when they authorized an investigation into future AMR. The investigation reported that by 2050, 10 million people could die from AMR-related illness per year. This staggering number implies a death every three seconds globally, mainly from diseases that were curable 30 years prior.
What can be done to stop AMR?
There are three main areas in which scientists are trying to tackle the issue of AMR.
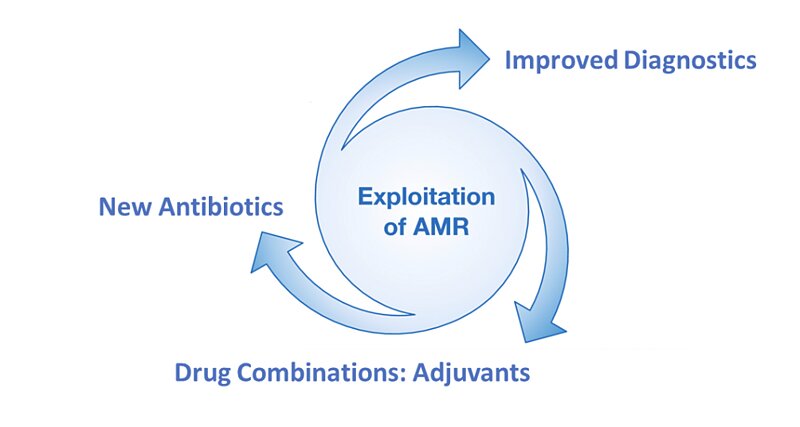 Figure 1: Scientific areas under active research to address the problem of AMR.
Figure 1: Scientific areas under active research to address the problem of AMR.
The first addresses the overuse or misuse of antimicrobial medicines, most notably antibiotics. All too often, antibiotics are given to patients that do not have bacterial disease or have such a mild bacterial infection that antibiotics are not warranted. This is thought to be the biggest driver of AMR in the modern era. Every single use of antibiotics places evolutionary pressure on bacterial microbes, which causes them to develop antibiotic resistance. Better and faster diagnostics promise the best solution to this problem, so that antibiotics are only given when they are truly needed.
The second, and arguably most obvious solution to AMR, is development of novel antimicrobial therapies. This, however, is difficult and very expensive, meaning production of new AMR-active therapies is rare. Despite the technical and financial barriers, antimicrobial therapies must continue to evolve if they are to maintain their effectiveness against ever-evolving pathogens.
The third possible solution is to enhance existing antimicrobial therapies by combining them with other drugs or ‘adjuvants’. This can mean that an antibiotic remains active against antibiotic resistant bacterial strains, or that it becomes active against a broader spectrum of bacterial targets. Clearly, there are huge benefits to be gained from this kind of approach. An everyday example is the widely used co-amoxiclav, which is a combination of amoxicillin and clavulanic acid. Clavulanic acid inhibits beta-lactamase, an enzyme produced by amoxicillin resistant bacteria.
Antibiotic resistance can be genotypic, meaning that a bacterium has evolved to be able to fight antibiotics; or antibiotic resistance can be structural, meaning that the bacteria form a physical barrier or ‘biofilm’, which protects at least some of the bacteria from the antibiotic. This kind of structural antibiotic resistance is often associated with relapse following antibiotic treatment, which is particularly prevalent in urinary tract infection (UTI) patients, of which there are around 150 million per year. When treated for a UTI, patients often report a relapse after a few days, which is caused because some bacteria survive the first course of antibiotics using a biofilm.
Combining an antibiotic with an adjuvant that breaks down biofilms
Wouldn’t it be great if an antibiotic could be combined with an adjuvant that breaks down biofilms, to address the issue of structural antibiotic resistance?
This is the idea which inspired a collaborative project lasting for over five years between the research group of Dr. Ashraf Zarkan, led by Dr. David Summers (Department of Genetics, University of Cambridge)1 and Cresset Discovery2. The Summers group identified the enzyme tryptophanase as being essential in the formation of biofilms, providing a target for small molecule adjuvant therapies. Working in collaboration with Cresset Discovery’s computational chemists, the Summers group were able to identify nine biofilm inhibitors that were shown to be effective against clinical isolates of pathogenic bacteria associated with UTI’s. These nine ‘hits’ also showed no cytotoxicity and suitable in silico ‘ADME’ (absorption, distribution, metabolism, and excretion) properties.
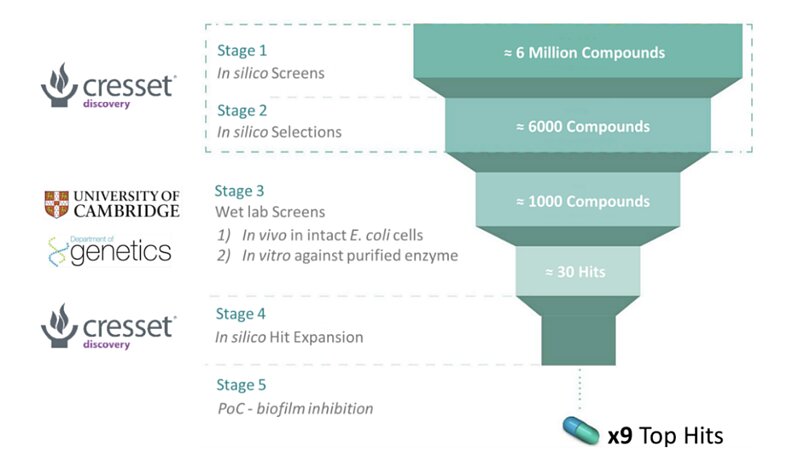 Figure 2: A breakdown of the collaborative work between the Summers group and Cresset Discovery.
Figure 2: A breakdown of the collaborative work between the Summers group and Cresset Discovery.
Cresset Discovery in silico methods offer several benefits to standard in vivo/in vitro assays for drug discovery, which are the driving force behind the collaboration. Screening millions of potential drug-like molecules is extremely time consuming and costly in a wet lab, while computational methods can offer similar conclusions in a much lower timeframe and at a fraction of the cost. Cresset’s task was to identify ‘hits’, i.e., molecules that are likely to show good activity against the tryptophanase target, with low toxicity (side effects), and that can easily enter the bacterial cell. To do this, the team used Cresset’s proprietary eXtended Electron Distribution (XED) force field3 to accurately map points of electrostatic extrema and hydrophobicity, on to six million candidate compounds from a chemical database. These points, known as field points, allow for rapid comparison of the important physical properties of the six million candidate compounds, and compounds that are already known to be active against the tryptophanase enzyme. This allowed Cresset Discovery scientists to quickly identify possible ‘lead’ compounds for further analysis, thus drastically increasing time and cost efficiency at later stages in the wet lab.
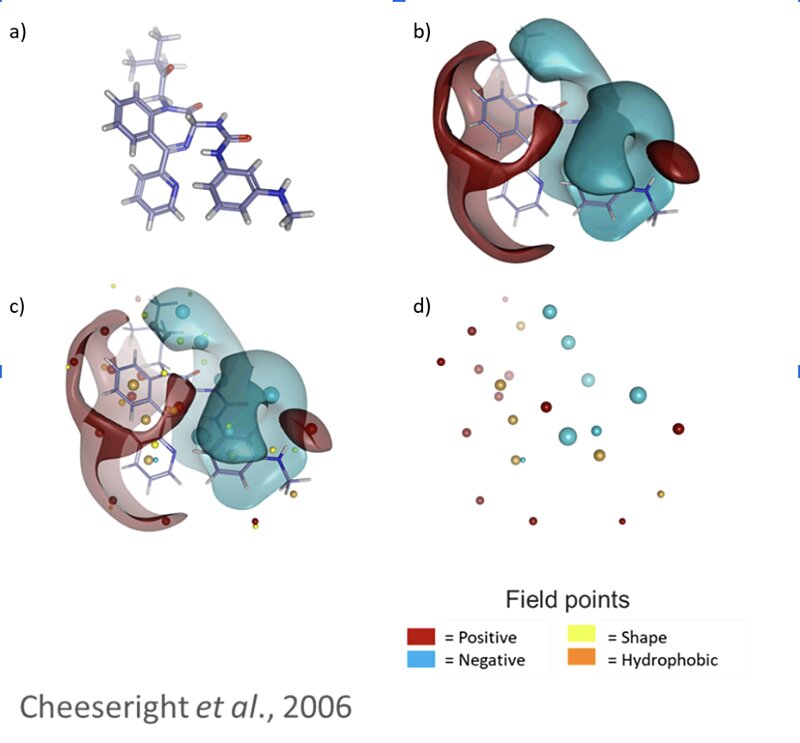 Figure 3: a) molecule in 3D space, b) Cresset’s XED forcefield can be used to accurately map the molecular interaction potential around the molecule, c) the surfaces contain lots of unnecessary information – we can assign field points to represent the extrema, d) this simplified representation can be used to rapidly screen against millions of compounds from a chemical database.
Figure 3: a) molecule in 3D space, b) Cresset’s XED forcefield can be used to accurately map the molecular interaction potential around the molecule, c) the surfaces contain lots of unnecessary information – we can assign field points to represent the extrema, d) this simplified representation can be used to rapidly screen against millions of compounds from a chemical database.
Optimizing research investment
The Cresset Discovery team continue to work closely with the Summers group as potential drug candidates are progressed towards clinical trials. We thank the Summers group and the University of Cambridge for their continued partnership.
To find out how to accelerate your discovery project, and optimize your research investment, contact enquiries@cresset-group.com or request a free confidential discussion.
Connect with Cresset Discovery.
References
- https://www.gen.cam.ac.uk/research-groups/research-groups/summers
- https://www.cresset-group.com/discovery/
- Vinter, J.G. Extended electron distributions applied to the molecular mechanics of some intermolecular interactions. J Computer-Aided Mol Des 8, 653 – 668 (1994)https://doi.org/10.1007/BF00124013